Please note that heatmap may take a few seconds to fully load, update, or download.
To change or update the data being displayed, use the Table of Results tab.
Reaction Biology (KiR 1)
This screen was performed through a collaboration between the Fox Chase Cancer Center and Reaction Biology Corporation. Details about the assay can be found in the original publication.
The data can also be explored here.
Citation: Anastassiadis, T., Deacon, S. W., Devarajan, K., Ma, H., & Peterson, J. R. (2011). Comprehensive assay of kinase catalytic activity reveals features of kinase inhibitor selectivity. Nature Biotechnology, 29(11), 1039-1045. https://doi.org/10.1038/nbt.2017
HMS LINCS
The Library of Integrated Network-based Cellular Signatures (LINCS) is an NIH-funded program at Harvard Medical School. This dataset uses the KINOMEscan Assay to profile kinases bound by inhibitors.
The resulting datasets can be accessed here.
Citation: Koleti, A., Terryn, R., Stathias, V., Chung, C., Cooper, D. J., Turner, J. P., . Schurer, S. C. (2017). Data Portal for the Library of Integrated Network-based Cellular Signatures (LINCS) program: integrated access to diverse large-scale cellular perturbation response data. Nucleic Acids Research. https://doi.org/10.1093/nar/gkx1063
GSK PKIS
A library of the GlaxoSmithKline (GSK) Published Kinase Inhibitor Set (PKIS) was assayed at the NIH using luciferase reporter enzymes. Details about the assay can be found in below publication.
The published datasets can be accessed by following the instructions found here.
Citation: Dranchak, P., MacArthur, R., Guha, R., Zuercher, W. J., Drewry, D. H., Auld, D. S., & Inglese, J. (2013). Profile of the GSK Published Protein Kinase Inhibitor Set Across ATP-Dependent and-Independent Luciferases: Implications for Reporter-Gene Assays. PLoS ONE, 8(3), e57888. https://doi.org/10.1371/journal.pone.0057888
EMD Millipore
Pairwise assays between purified recombinant human kinases and a collection of small molecule inhibitors were carried out by the EMD Millipore Corporation (now known as Millipore Sigma) using a filter binding radioactive ATP transferase assay.
Raw data and details on the recombinant kinases and inhibitors used can be found in the supplemental files of the original publication here.
Citation: Gao, Y., Davies, S. P., Augustin, M., Woodward, A., Patel, U. A., Kovelman, R., & Harvey, K. J. (2013). A broad activity screen in support of a chemogenomic map for kinase signalling research and drug discovery. Biochemical Journal, 451(2), 313-328. https://doi.org/10.1042/BJ20121418
Submitting a New Dataset
If there is a dataset or compound screen that you would like to see included in this tool, please contact the authors of the publication (link in sidebar).
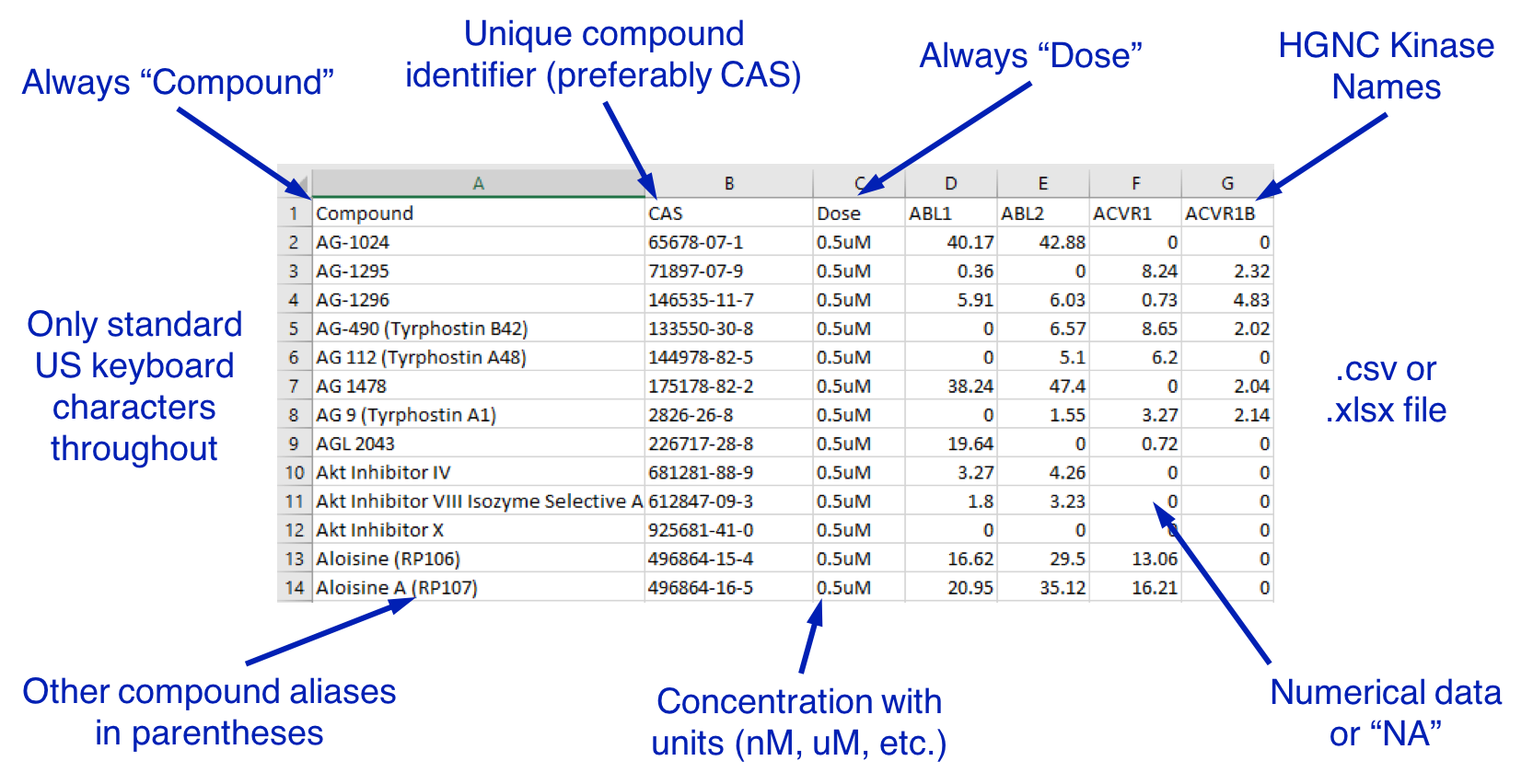
File Guidelines for Submitting a Dataset:
- File should be in (.csv) format. Excel 2007+ (.xlsx) format is also acceptable, but DO NOT use (.xls) as this may lead to truncation and loss of data.
- Only characters on a standard US keyboard should be used throughout the file (e.g. use "u" in place of a greek mu).
-
First column should be labeled "Compound" and contain compound names, with aliases in parentheses.Ex: Dorsomorphin (BML-275, Compound C)
- Second column should be a unique identifier, preferably CAS numbers. Columnn header should be "CAS" or appropriate identifier name.
- Third column should be labeled "Dose" and contain the dose and unit (no spaces) of the compound tested.
- All subsequent column headers should be standardized kinase names (see below for kinase naming conventions).
- All entries in the table must be numerical, percent-of-control inhibition values between 0 and 100 (100 = total inhibition, 0 = no inhibition compared to control), or "NA" for missing values.
- Include the source citation / publication for the data or otherwise demonstrate data access permission.
Kinase Naming Conventions:
-
Use the HGNC gene names (in all caps) for all protein names.
- Do not include any whitespace characters (spaces, tabs, newlines, etc) in kinase names.
-
Indicate mutations in parentheses as "(X###Y)". Separate additional mutations with a comma, and indicate deleted regions with "del".Ex: EGFR(L747-E749del,A750P).
-
Include complexed or bound proteins with their HGNC gene names, separated by a "+".Ex: CDK2+CCNA2
-
Denote activated (cleaved, etc.) forms without distinct HGNC names with "(a)".Ex: CDK5+CDK5R1(a)
- Denote inactivated or inhibited forms with "(i)".
- Denote phosphorylated proteins with "(p)".
-
Denote non-mutant variants (e.g. splice variants) without distinct HGNC names with "(v2)", "(v3)", etc., but do not use "(v1)" for the most common variant.Ex: LYN, LYN(v2)
-
Denote multiple domains of the same kinase screened separately with "(dom1)", "(dom2)", etc., numbered from N-terminus to the C-terminus.Ex: JAK1(dom1), JAK1(dom2)